
Dina Schneidman
Associate professor
dina.schneidman {at} mail.huji.ac.il
- Integrative structure modeling of protein-protein interactions and large macromolecular assemblies
- Structure modeling and design of adaptive immune response interactions, including antibody-antigen and peptide-MHC-TCR systems
- Deep learning methods for protein structure modeling, specificity, and design
- Small angle x-ray scattering (SAXS)
- Crosslinking mass spectrometry

Merav Braitbard
PhD student
merav.braitbard {at} mail.huji.ac.il
- Integrative structure modeling of RSC assembly
- Specificity of peptide-SH3 interactions
- Modeling large macromolecular complexes with crosslinking mass spectrometry datasets

Tomer Cohen
PhD student
tomer.cohen13 {at} mail.huji.ac.il
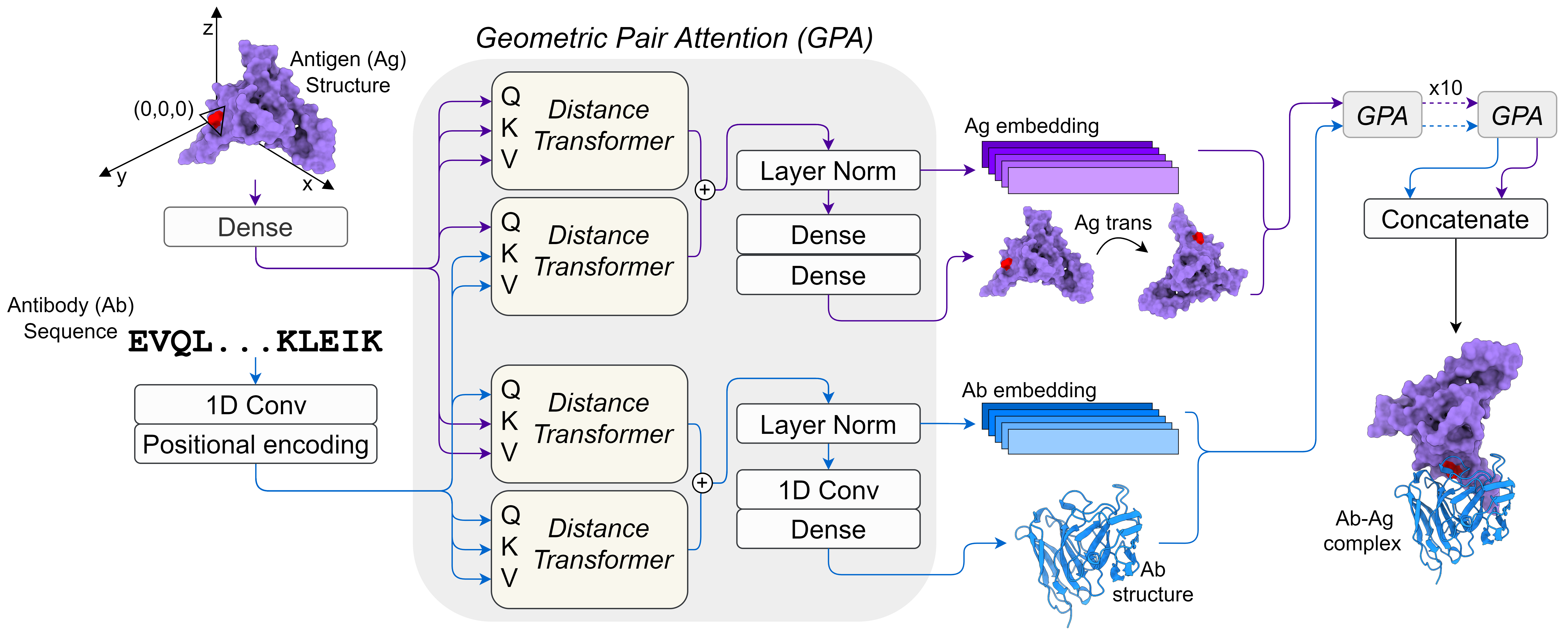
- Methods for nanobody/antibody modeling by deep learning
- Antibody-antigen structure prediction
- Antibody design

Ben Shor
PhD student
ben.shor {at} mail.huji.ac.il
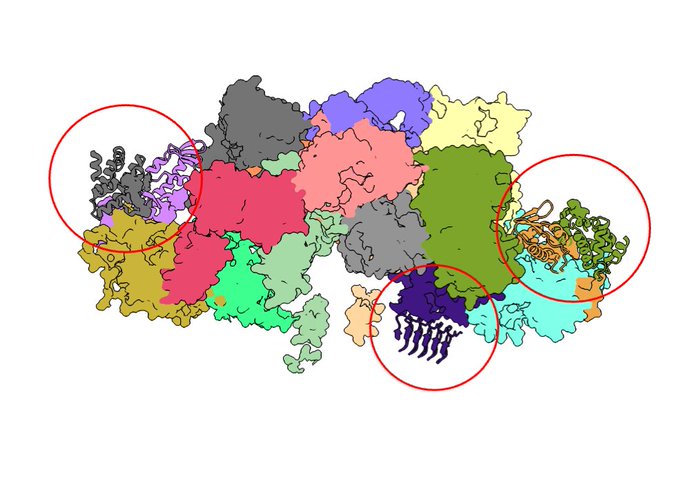
- Assembly of large macromolecular complexes

Gal Passi
MSc student
gal.passi {at} mail.huji.ac.il
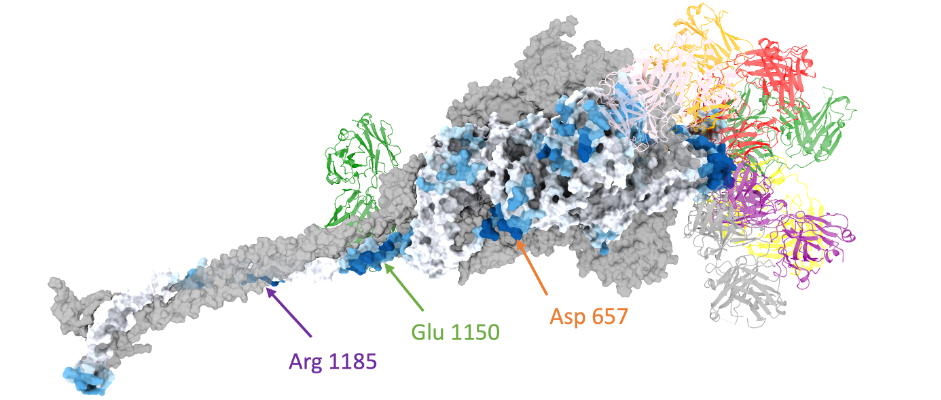
- Interpreting cancer mutations with deep learning models

Tanya Hochner
MSc student
tanya.hochner {at} mail.huji.ac.il
Deep learning models for predicting structure and specificity of peptide-MHC-TCR systems

Alon Aronson
MSc student
alon.aronson {at} mail.huji.ac.il
Deep learning models for predicting structure and specificity of peptide-MHC-TCR systems

Meitar Sela
MSc student
meitar.sela {at} mail.huji.ac.il
Deep learning models for predicting rhodopsin spectra maxima

Adi Weshler
PhD student (joint with Prof. Michal Linial)
{at} mail.huji.ac.il

Amir Weinfeld
MSc student (joint with Prof. Jérôme Tubiana)
amir.weinfeld {at} mail.huji.ac.il
- Predicting peptide-SH3 specificity with deep learning
- Predicting anti-drug-antibody (ADA) potential

Danielle Zaccai
Computational Biology project
danielle.zaccai {at} mail.huji.ac.il
Upresolution of single molecule data
Alumni
- Prediction of protein binding sites
- Antibody epitope prediction
- Protein design
- Development of novel deep learning architectures for protein structures

Edan Patt
MSc student
edan.patt {at} mail.huji.ac.il
- Prediction of magnesium sites in RNA structures
- Modeling RNA dynamics with SAXS profiles

Matan Halfon
MSc student
matan.halfon {at} mail.huji.ac.il
Scoring functions for protein-protein docking.

Shon Cohen
MSc student
shon.cohen {at} mail.huji.ac.il
Scoring functions for cross-linking MS data.

Nurit Meyer
MSc student
nurit.meyer {at} mail.huji.ac.il
Modeling Hsp33-substrate interactions

Amichai Holzer
MSc student
amichai.holzer {at} mail.huji.ac.il
Amichai is developing a method for fitting structural fragments into cryo EM density maps.
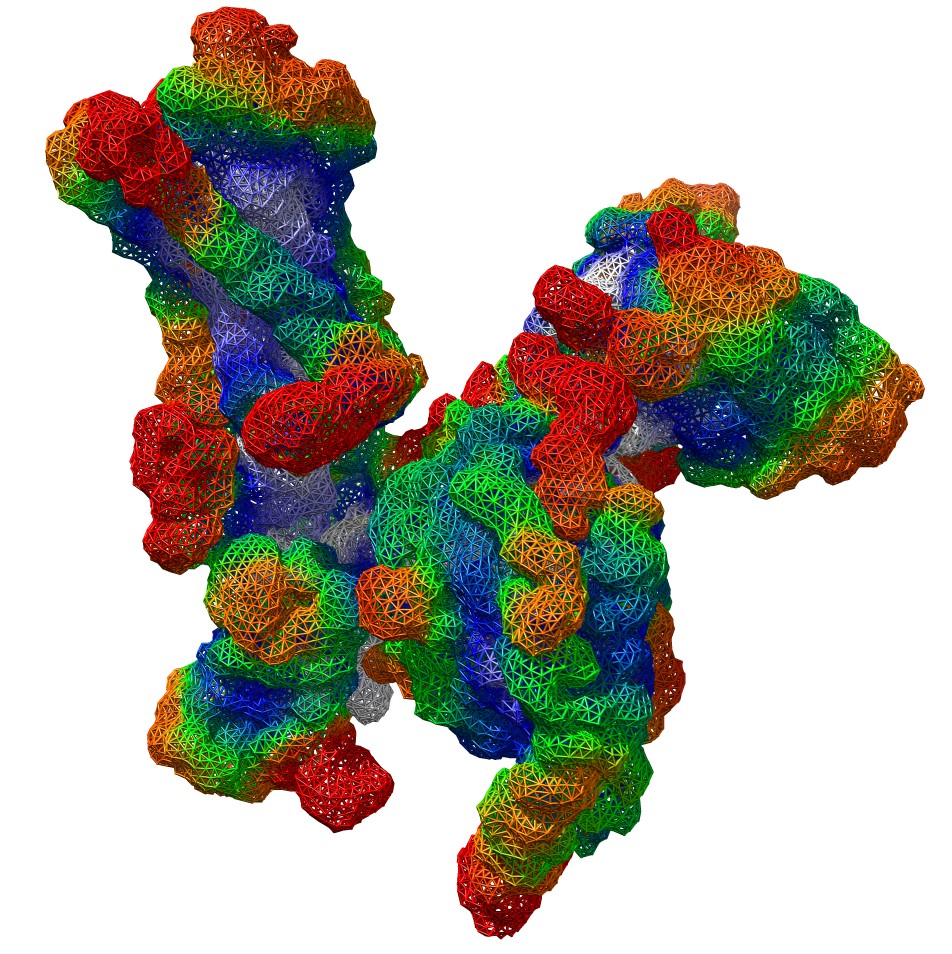
A surface mesh of the density map of mu opioid receptor-gi protein complex (EMDB:7868) colored by Heat Kernel Signature.

A surface mesh of the density map of mu opioid receptor-gi protein complex (EMDB:7868) colored by Heat Kernel Signature.
Lirane Bitton
MSc student
lirane.bitton {at} mail.huji.ac.il
End-to-end deep learning of 3D structures for prediction of HLA class II epitopes.
Classification of nanobodies by affinity using machine learning.
